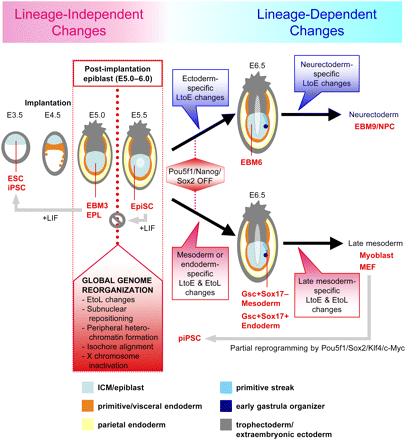
A model: Lineage-independent and lineage-dependent replication-timing changes before and after the post-implantation epiblast stage, respectively. Schematic diagrams of mouse embryos at different stages of embryogenesis are shown along with cell types that model different tissues and stages analyzed in this study (red). We propose that lineage-independent EtoL changes are completed by the post-implantation epiblast stage (E5.0–E6.0), prior to germ layer specification and down-regulation of key pluripotency transcription factors (POU5F1/NANOG/SOX2). These changes are accompanied by several events listed, which collectively represent a form of genome reorganization (“autosomal Lyonization”) that is coincident with EpiSCs having lost the ability to easily revert back to the ESC state. Lineage-dependent EtoL and LtoE changes occur continuously after the late epiblast stage to create cell-type-specific profiles. piPSCs are trapped at an epigenetic state that has not yet reprogrammed this epiblast stage genome reorganization.











