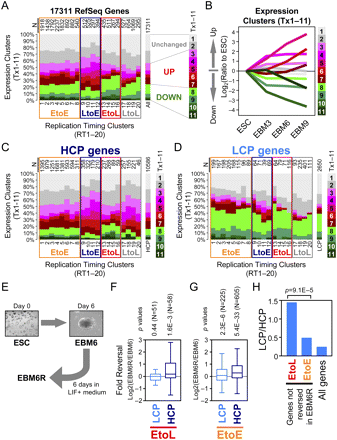
Relationship between replication-timing changes, transcription changes, and their reversibility (A) Cross-tabulation of K-means clusters of replication timing and transcription. All 17,311 RefSeq genes present on the gene expression array were analyzed. Each of the 20 replication-timing clusters, RT1–20 (Fig. 2F), were presented as a stacked bar consisting of the 11 expression clusters, Tx1–11 (Fig. 1C). N, number of genes in RT1–20. (B) Line chart showing the kinetics of transcription changes of Tx1–11 during differentiation. Log2 transformed ratios to ESC levels are plotted. (C,D) The same cross-tabulation graph as in A, for 10,586 HCP genes (C) and 2650 LCP genes (D). HCP and LCP classification is based on Mikkelsen et al. (2007). (E) A scheme for the reversal of differentiation. EBM6R is a condition in which EBM6 cells were placed back into ESC medium containing LIF and cultured for 6 d. (F,G) Box plots showing the reversibility of different classes of genes. Log2 transformed expression level in EBM6R relative to EBM6 is plotted. Twofold down-regulated genes during the ESC–EBM6 transition were selected from the down-regulated expression clusters, Tx8–11. Among them, genes in the EtoL clusters (RT13–16) are presented in F, while genes in the EtoE clusters (RT1–8) are presented in G. P-values were obtained from a two-tailed t-test for comparison of two paired groups (i.e., EBM6R vs. EBM6). (H) LCP is overrepresented in EtoL genes that are not reversed in EBM6R. Twofold down-regulated EtoL and EtoE genes that did not show reversal (i.e., EBM6R/EBM6 < 1) were calculated for their LCP-to-HCP ratio. Note that there are four times as many HCPs as LCPs in the genome (=All genes). Numbers of genes in each category are 49 (EtoL) and 313 (EtoE). A P-value was obtained from a χ2 test.











