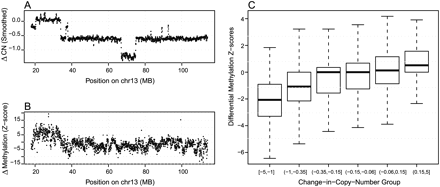
Figure 7.
Effects of copy number changes on differential methylation detection. (A) Differential methylation Z-score for between LNCaP and PrEC cells, using MBD-SF-seq, for human chromosome 13. (B) Smoothed Affymetrix SNP 6.0 array data showing corresponding changes in copy number. (C) Genome-wide distributions of Z-scores, stratified by the change-in-copy-number status of the corresponding regions.











