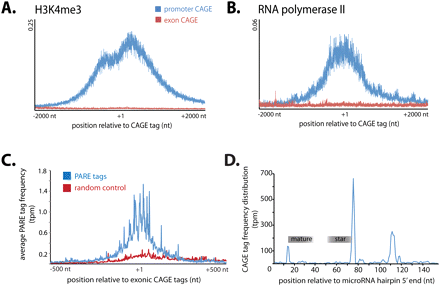
Frequency distribution for modified chromatin immunoprecipitated tags relative to CAGE sites associated with gene promoters and exons. (A,B) The average frequency of DNA tags immunoprecipitated with antibodies to H3K4me3 (A) and RNA polymerase II (B) within a 4-kb window centered on CAGE tags mapping to promoters (blue) and coding exons (red) (Wang et al. 2008). (C) Frequency distribution of PARE tags (blue) within a 1000-nt window centered on exonic CAGE tags (indicated at +1). A random control showing PARE tags frequency distribution across random sites in matched exons shown (red). (D) Frequency of CAGE tags mapping across microRNA hairpin with mature and star sequences indicated.











