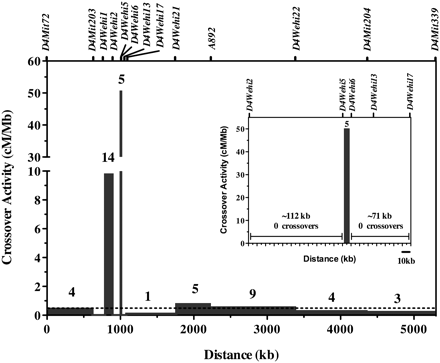
Distribution of crossover breakpoints within the NOD.B6Idd11D congenic interval. Seven-hundred-twenty-three F2 pups (1446 meiosis events) were derived from intercrossing NOD.B6Idd11D F1 mice and were genotyped for markers within the NOD.B6Idd11D congenic interval. The crossover activity was calculated by dividing the frequency of crossovers in each interval by its length. For example, five pups had a recombination between D4Wehi5 and D4Wehi6, giving rise to a genetic distance of 0.345 cM across this 6859-bp interval, which is ∼50 cM Mb-1. Bars, crossover frequency in cM/Mb in each interval. Above each bar, the number of crossovers observed in that marker interval is shown. The dashed line indicates the average crossover activity for the mouse genome: ∼0.5cM/Mb (Shiroishi et al. 1995). Genetic markers are indicated by ticks above the plot. The inset displays an enlarged view of the interval between D4Wehi2 and D4Wehi17.











