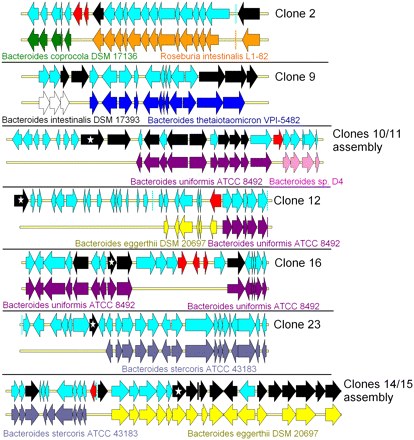
Evidence of horizontal gene transfers (HGTs) in human gut metagenomic sequences. HGTs were identified when rupture was observed in gene synteny between the genes present in the metagenomic DNA fragments and their best BLASTP hits issued from sequenced genomes. For each clone, the first line represents the clone metagenomic sequence, and the second line represents the genome part in synteny with it. Each arrow represents a gene. (Red arrows) Genes encoding putative transposases or integrases; (black arrows) CAZy-encoding genes; (stars within black arrows) genes encoding the CAZymes involved in the activity detected in the primary screens, as proven by transposon insertion in the fosmid inserts.











