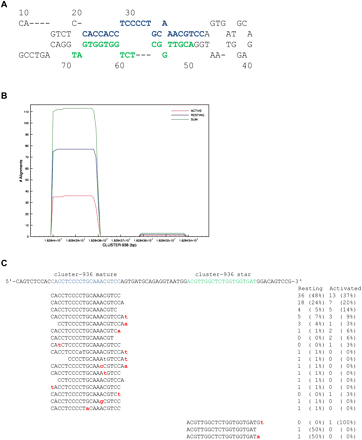
Novel miRNA genes identified in NK cells. (A) Predicted secondary fold structure of a novel miRNA precursor hairpin cluster-936 (mmu-mir-1306) (16:18284332..18284410) identified in Illumina GA–sequenced NK cell small RNAs (predicted ΔG = −30.7). Mature miRNA is noted in blue, minor star species in green. (B) Read alignment coverage map (RefCov) of novel miRNA gene demonstrating the specific alignment of sequences to the mature miRNA sequence. Shown is the nucleotide position of the precursor (x-axis) and the number of reads aligning to each nucleotide position (y-axis). Coverage models for total (sum = resting and activated), resting, and activated NK libraries are shown. (C) IsomiR analysis of novel miRNA with mature sequence noted in blue, and the read counts and corresponding percentage contribution to the total number of reads aligning to this novel hairpin.











