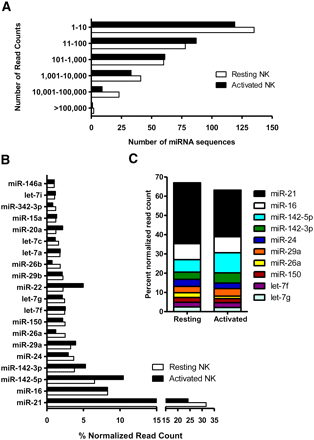
Known miRNAs sequenced in resting and activated NK cells using the Illumina GA. (A) Distribution of miRNA sequences in resting and activated NK cells based on the total read counts. Mature miRNA sequences are categorized by the number of read counts corresponding to each individual miRNA. (B) Top 20 known miRNAs expressed in resting and activated NK cells. The percent contribution of each miRNA sequence to the total pool of known miRNA sequences was calculated by dividing individual miRNA read counts by the total number of known miRNA sequence reads in that cDNA library. (C) Top 10 known miRNAs constitute ∼65% of total miRNA reads in NK cells.











