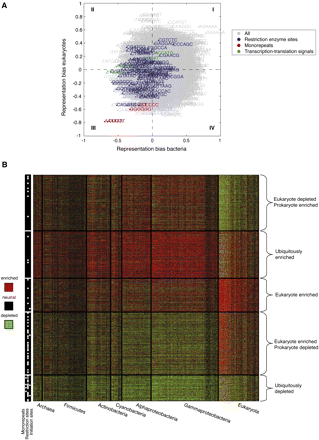
A global view of the additional information encoded within protein-coding sequences. (A) Comparison of short sequence enrichments in coding regions between bacteria and eukaryotes. For each short sequence, shown is its overall enrichment in bacteria (x-axis) and eukaryotes (y-axis), where the overall enrichment in each of the two phyla groups is taken to be the difference between the fraction of phyla species in which the sequence is enriched in the real versus randomized genome (at P < 0.05) and the fraction of species within that phyla is which it is depleted. (Red) Sequences that correspond to mononucleotide repeats; (blue) sequences that correspond to bacterial restriction enzyme sites; (green) sequences that correspond to bacterial transcription and translation initiation sites. (B) A clustering representation of the log-ratio coding region enrichment of all 6-mers across all 363 organisms whose coding regions exceed 2 Mbp. Rows, 6-mers; columns, organisms. The data were clustered using k-means clustering (k = 5) of a reduced matrix of representation bias in archaea, bacteria, and eukaryotes. White marks on the left bar indicate three specific sequence families: mononucleotide repeats (left column), restriction enzyme sites (central column), and transcription and translation initiation sites (right column). The organisms are arranged in phyla groups and are shown on the bottom. Red and green denote enrichment and depletion (P < 0.05) above that expected in randomized genomes, respectively.











