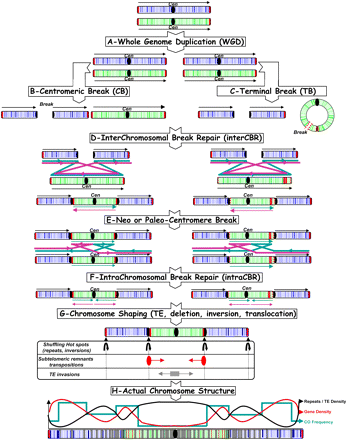
Model for grass chromosome evolution and shuffling. The model begins with WGD (A), followed by centromeric breaks (B) or terminal breaks (C), interchromosomal break repair (D), and intra-CBR mechanisms (E, F), respectively, between nonhomologous (A) and homologous chromosomes (E) to explain the observed NCF and CI pattern (G) as well as repeat/TE, gene, and CO distribution in rice, maize, sorghum, and Brachypodium genomes, resulting from their paleo-history from their common AGK (H). Colored arrows represent the different alternative orientation of the double-strand break repair to explain the actual syntenic chromosome order and orientation observed among rice, Brachypodium, sorghum, maize.











