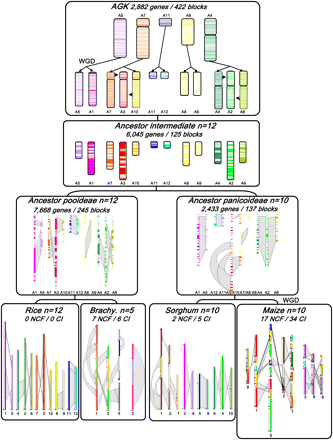
Ancestral grass karyotype reconstruction. The monocot (rice, Brachypodium, sorghum, maize) chromosomes are represented with color codes to illustrate the evolution of segments from a common ancestor with five protochromosomes (named according to the rice nomenclature A5, A4, A7, A8, A11). The current structure of the four genomes is represented at the bottom of the figure, with the seven ancestral duplications highlighted with gray boxes. Large segmental inversions are indicated with red arrows in Brachypodium, sorghum, and maize genomes according to the synteny, with rice used as reference genome. The ancestor intermediate (n = 12 for Pooideae, A1 to A12; and n = 10 for Panicoideae, A1 to A10) is illustrated by blocks of reordered genes, with the chromosome and duplication five-color code described above. The AGK (top), structured in seven blocks of five protochromosomes, went through a WGD and two chromosome fusions and fissions (A3 = A7 + A10; A2 = A4 + A6) to reach the n = 12 ancestor intermediate. Polyploidization events are indicated as WGD in the figure. The number of reordered genes, gene blocks, NCF, and CI events are indicated on the depicted modern and ancestral genomes.











