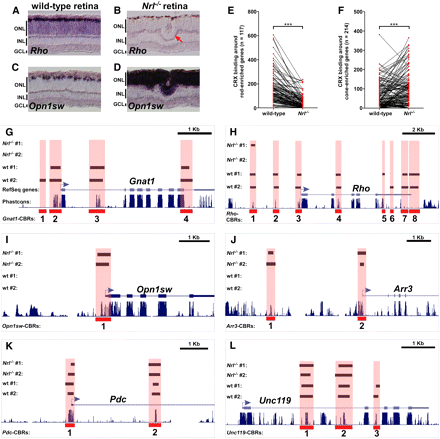
Rods and cones have both shared and cell type–specific CRX-bound regions. (A–D) In situ hybridization pattern of rod-specific rhodopsin (Rho) on wild-type (A) and Nrl−/− (B) retinas, and cone-specific blue opsin (Opn1sw) on wild-type (C) and Nrl−/− (D) retinas. In the wild-type retina, Rho is expressed in the majority of cells in the ONL, whereas Opn1sw is only expressed in a small subset of cells at the outer edge of the ONL. The Nrl−/− retina shows the converse pattern: Rho is completely absent, whereas Opn1sw is strongly expressed throughout the entire ONL. Rosette formation in common in the ONL of Nrl−/− retinas (red arrow in B). (E) CRX binding around rod-enriched genes in wild-type and Nrl−/− retinas. Each pair of red dots connected by a black line represents a single rod-enriched gene. The y-axis indicates the number of sequence reads within all CBRs assigned to that gene. There is a marked decrease in the number of assigned sequence reads for most rod genes in the Nrl−/− retina relative to wild-type. ***P < 0.0001, paired Student's t-test. (F) CRX binding around cone-enriched genes in wild-type and Nrl−/− retinas. In this case, there is an overall increase in CRX binding around cone genes in Nrl−/− retinas compared with wild-type. ***P < 0.0001, paired Student's t-test. Gnat1 (G) and Rho (H), both rod-specific genes, show a near absence of CBRs in the Nrl−/− retina. Opn1sw (I) and Arr3 (J), both cone-specific genes, show prominent CBRs in the Nrl−/− retina but not in wild-type. Pdc (K) and Unc119 (L) are expressed at similar levels in both rods and cones. They show similar levels of CRX binding in both wild-type and Nrl−/− retinas.











