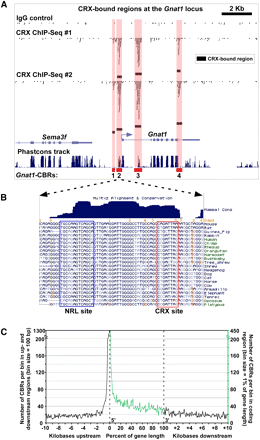
Distribution of CRX-bound regions around photoreceptor genes. (A) Pattern of CRX-bound regions around Gnat1 that encodes rod alpha-transducin, a component of the phototransduction cascade. Sequence reads derived from two ChIP-seq replicates using an anti-CRX antibody (“CRX ChIP-seq #1” and “CRX ChIP-seq #2”) or an IgG control (“IgG control”) are shown along with the corresponding CBRs. Also shown is the “Phastcons track,” which indicates the pattern of phlyogenetic conservation across the region (Siepel et al. 2005). In this and subsequent figures, CBRs are numbered from 5′ to 3′ with respect to the transcription start site of the gene with which they are associated. (B) Sequence-level view of a portion of Gnat1-CBR2. Note the presence of phylogenetically conserved CRX and NRL binding sites within this region. Additional conserved motifs are also evident, but their binding factors are currently unknown. (C) Distribution of CBRs around mouse genes. This graph shows the density of CBRs over the length of all mouse genes, as well as in the first 10 kb upstream of and downstream from all genes. Note that the location of CBRs within genes is given as percentage of gene length.











