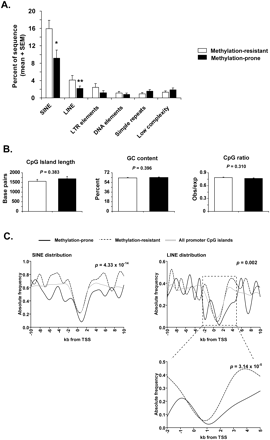
Distribution of repetitive elements in methylation-prone versus methylation-resistant genes. (A) The abundance of repetitive elements of different classes was determined for the 4-kb sequence window centered in the TSS of 36 methylation-resistant (white) and 36 methylation-prone (black) genes. Retrotranposons of the SINE and LINE classes were found to be depleted in methylation-prone genes. *P < 0.02; **P < 0.12 (Student's t-test). (B) Average length, GC content, and CpG ratio of CpG islands were not significantly different between methylation-prone and methylation-resistant genes. Error bars represent SEM. (C) Abundance of SINE and LINE retrotransposons in the 20-kb sequence window centered in the TSS of 36 methylation-prone and 36 methylation-resistant genes. The abundance of SINE and LINE retrotransposons in all promoter CpG islands in the human genome is shown in gray. Note that the depletion of LINE retrotransposons is more significant in the −2-kb to +5-kb sequence window.











