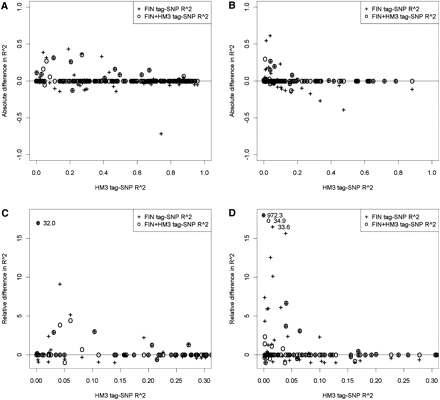
Comparison of the correlations (R2) between CNP signal and tag-SNP genotype in the H2000 data. Three tag-SNPs were tested for each CNP; the tag-SNP with the highest R2 value between SNP genotypes and CNP signal in HM3 data, FIN data, and FIN + HM3 data. Data is plotted here separately for biallelic CNPs of common (MAF ≥ 5%, N = 192) (A,C) and low-frequency (MAF between 0.5% and 5%, N = 89) (B,D). In top plots, FIN + HM3 tag-SNP and FIN tag-SNP sets are compared with HM3 tag-SNPs by plotting the FIN + HM3 and FIN differences with the HM3 tag-SNP against the HM3 tag-SNP R2 value, all from the H2000 correlation analysis. A negative difference indicates that the HM3 tag-SNP has a stronger correlation with the CNP signal in the H2000 data. If the difference is positive, then the correlation is better when the CNP tag-SNP for H2000 is selected using FIN + HM3 or FIN data. At bottom we show the relative differences in the R2 values ([FIN or FIN + HM3 R2–HM3 R2]/HM3 R2), plotted against the HM3 tag-SNP R2 value. We focus on the CNPs where the HM3 tag-SNP R2 value is <0.3, since this is the area where the highest relative differences are possible. One observation in C and three in D are of significantly larger values than the rest of the data; these are shown in the top left region of the plot with their corresponding values next to them. (HM3) HapMap3 reference set; (FIN) Finnish HapMap3; (FIN + HM3) combined reference.











