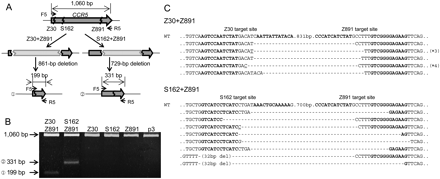
Use of two ZFNs for targeted genome deletions within the CCR5 locus. (A) Schematic representation of two different deletion events within the CCR5 locus. F5 and R5 are PCR primers (arrows) used for the detection of genome deletion events. (B) PCR products validating ZFN-induced genomic deletions. Approximate sizes of PCR products corresponding to deletion events (199 bp and 331 bp) and to the intact wild-type sequence (1060 bp) are indicated as predicted in A. p3 is the empty plasmid used as a negative control. (C) DNA sequences of PCR products corresponding to deletion events. Symbols are as in Figure 1.











