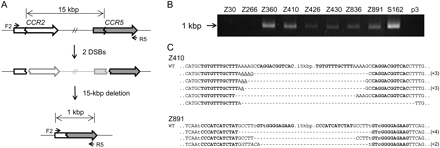
ZFN-induced genome deletions at the CCR2 and CCR5 loci. (A) Schematic representation of ZFN-mediated genome deletions. Zigzag lines indicate ZFN target sites. F2 and R5 are PCR primers (arrows) used for the detection of genome deletion events. (B) PCR products corresponding to the 15-kbp genomic DNA deletions in cells treated with ZFNs. p3 is the empty plasmid used as a negative control. (C) DNA sequences of PCR products. PCR products were cloned and sequenced. ZFN target sites are shown in boldface letters. Microhomologies are underlined and inserted bases are shown in italics. Dashes indicate deleted bases. Nonconserved bases at the CCR2 and CCR5 loci are shown in lowercase letters. In cases in which a deletion sequence was detected more than once, the number of occurrences is shown in parentheses. (WT) Wild-type DNA sequence.











