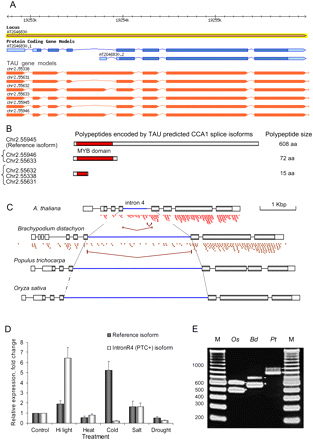
Intron retention and novel splice junction events in the CCA1 locus. (A) Empirical CCA1 gene models (orange) generated by the TAU tool using RNA-seq data. (B) Predicted polypeptides are shown schematically with the DNA binding MYB domain shown by a red box. (C) Gene models of homologous CCA1/LHY loci in A. thaliana, Oryza sativa, Brachypodium distachyon, and Populus trichocarpa. cDNA microread coverage is shown for Arabidopsis and Brachypodium. SJs of intron 4 and 4a splicing in Arabidopsis and Brachypodium are marked by brown broken lines. (D) Quantification of the IntronR4 event by qRT-PCR under different abiotic stress conditions. Lanes labeled Hi Light, Heat, Cold, Salt, and Drought correspond to high light, heat, cold, salt, and dehydration treatments, respectively. Relative expression was estimated using −ΔΔCt method (Livak and Schmittgen 2001) and EF-1-ALPHA mRNA as an internal housekeeping gene control. (E) RT-PCR confirmation of CCA1 IntronR4 in rice, poplar, and Brachypodium. IntronR4-specific primers were designed as described for A. thaliana (as shown in panel B). RT-PCR products corresponding to the retained intron 4 (if downstream intron 5 is spliced) are denoted by an asterisk (*); pre-mRNAs are indicated by a dash (–). Sanger sequencing of gel-purified amplified DNA fragments confirmed the sequence of all RT-PCR products. The predicted fragment sizes are 492, 573, and 782 bp for rice (Os, Oryza sativa, ssp. Japonica, locus ID: LOC_Os08g06110), Brachypodium (Bd, Brachypodium distachyon, locus ID: Bradi3g16510), and poplar (Pt, Populus trichocarpa, locus ID: estExt_Genewise1_v1.C_LG_XIV1950), respectively.











