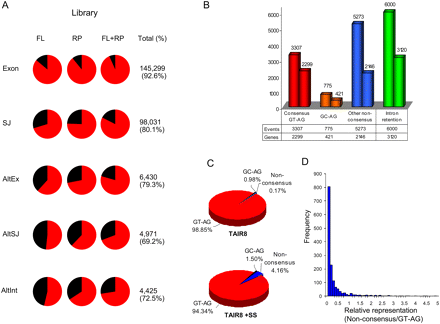
Survey of constitutive and alternative splicing in Arabidopsis. (A) Detection of annotated gene features and alternative splicing events by RNAseq. Annotated gene features (Exon, SJ) and alternative splicing events, including alternative splicing at both acceptor and donor splice sites (AltEx), an alternative splice junction (AltSJ) and alternative intronic sequences (AltInt) were identified by aligning RNA-seq microreads as described in the Supplemental material. Pie charts depict the proportions of the annotated features in full-length (FL), randomly primed (RP), and combined (FL + RP) cDNA libraries detected by at least one perfect match RNA-seq read. Total (%) indicates the total number and percentage of annotated features detected in the combined (FL + RP) data. (B) Distribution of novel splicing events among annotated genes. The histogram depicts the numbers of novel alternative splicing events and alternatively spliced genes containing consensus GT-AG and other nonconsensus intron splice signal dinucleotides and introns retention events within TAIR8-annotated genes. (C) Pie charts depict the proportions of consensus and nonconsensus intron terminal dinucleotide classes among annotated TAIR8 introns (TAIR8; upper panel) and the combined TAIR8 + supersplat-predicted introns (TAIR + SS; lower panel). (D) The histogram depicts the relative representation of consensus and nonconsensus splice junctions as a frequency distribution. The relative representation was calculated (the average number of reads spanning nonconsensus splice junctions/the average number of reads spanning constitutive consensus splice junctions in the same gene) for 1539 genes that contain both consensus and nonconsensus introns.











