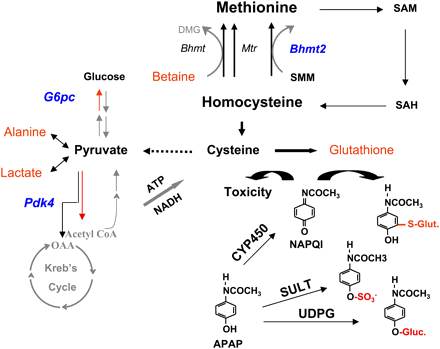
Metabolic pathways affecting sensitivity to acetaminophen (APAP)-induced hepatotoxicity. SJL/J mice that are uniquely resistant to APAP-induced liver toxicity had four endogenous metabolites (red text) with altered abundances, and three enzymes (blue text) had altered gene expression after drug exposure. The metabolic pathways encompassing these enzymes and endogenous metabolites are diagrammed. Cysteine biosynthesis is the rate-limiting step for glutathione (GSH) production. Alterations in Bhmt2 mRNA and betaine abundance that favor cysteine production would contribute to resistance to APAP-induced liver injury. Cysteine can be converted to pyruvate, which is in equilibrium with alanine and lactate. An increase in cysteine biosynthesis would explain why alanine and lactate were increased in the resistant mice. Reduced G6pc and Pdk4 will inhibit gluconeogenesis and favor pyruvate breakdown to increase cellular reducing capacity. These metabolic alterations might help SJL/J mice resist an APAP-induced toxic challenge. All three homocysteine methylation enzymes (Bhmt, Mtr, Bhmt2) are depicted in the figure. BHMT2 is an S-methyl-methionine (SMM)-specific methyltransferase and cannot use betaine as the methyl donor. Betaine is a required substrate for BHMT-catalyzed remethylation of homocysteine to form methionine. Sequential steps for SMM- and S-adenosyl methionine (SAM)-dependent methionine/GSH biosynthesis pathways are shown. APAP clearance by sulfation (SULT) or glucuronidation (UDPG), and cytochrome P450 (CYP450) mediated generation of NAPQI are depicted in the figure. Excess NAPQI would induce toxicity.











