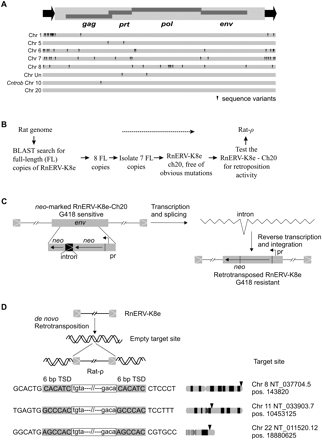
Identification of an autonomous RnERV-K8e element. (A) Genomic organization of a full-length, autonomous RnERV-K8e element, with the 429-bp-long LTRs (black arrows) flanking a 8332-bp sequence containing four complete open reading frames (ORFs) encoding the retroviral gag, prt, pol, and env genes. Eight full-length copies of the RnERV-K8e were found in the rat genome (assembly RGSC v3.1, see Supplemental material). The dark triangles indicate sequence variants that likely render the RnERV-K8e nonfunctional (see also Supplemental Table S2). The copy inserted into Cntrob contains a single point mutation disrupting the gag gene. The copy found on chromosome 20 (of the BN strain) contains no obvious mutations and was tested for functionality in a retrotransposition assay. (B) Flowchart of the identification of an autonomous copy of the RnERV-K8e family. (C) A reporter neo-cassette was incorporated into the env gene of the RnERV-K8e-Ch20 element with the neo gene and a promoter (pr) in reverse orientation. The marker cassette is inactive due to the insertion of an intron. Retrotransposition is assayed by the activation of the neo gene. The estimated retrotransposition frequency is 5 × 105–2 × 106, one and two orders-of-magnitude lower than those of IAP (Dewannieux et al. 2004) and MusD elements (Ribet et al. 2004), respectively. (D) Examples of de novo retrotransposition events of the RnERV-K8e-Ch20 element isolated from genomic DNA of G418 resistant HeLa colonies. Insertion junctions are depicted with endogenous sequences in uppercase and proviral sequences in lowercase. The retrovirus is flanked by canonical 6-bp target site duplication sequences (TSD, shaded). Arrows mark the approximate chromosomal locations of the de novo retrotransposition events. Exact positions of the target sites are shown on the right.











