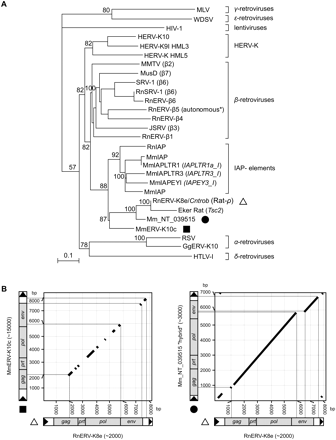
Phylogenetic characterization of the RnERV-K8e family. (A) Phylogeny of retroviruses based on the neighbor-joining method using highly conserved nucleotide sequence regions of the pol proteins (Baillie et al. 2004) rooted using a non-LTR retrotransposon. Percent bootstrap values, obtained from 1000 replicates, are indicated. The ERV families are indicated on the right. MLV, Moloney murine leukemia virus; WDSV, walleye dermal sarcoma virus; HIV-1, human immunodeficiency virus type 1; HERV-K, human endogenous retrovirus type K; MMTV, mouse mammary tumor virus; MusD, Mus musculus type D element; SRV-1, Simian retrovirus type D; RnSRV-1, SRV-1 from Rattus norvegicus (Rn); RnERVβ(1-6), β(1-6)-type ERVs from Rattus norvegicus; JSRV, jaagsiekte sheep retrovirus; IAP elements, Intracisternal-A particle elements from Mus musculus (Mm), from Rattus norvegicus (Rn), or Mesocricetus auratus (Ma); RnERV-K8e (Rat-ρ), endogenous retrovirus identified in this study (indicated by a triangle); Eker rat, a nonautonomous element inserted into the Tsc2 gene; Mm_NT_039515 (indicated by a filled circle); MmERV-K10c (indicated by a filled square), ERV-K-type element from Mus musculus; RSV, Rous sarcoma virus; GgERV-K10, ERV-K-type element from Gallus gallus; HTLV-1, human T-cell leukemia virus type-1. All sequences are accessible from GenBank or Rebpase (see Supplemental material). Scale bar indicates nucleotide divergence. (B) Dot plots of RnERV-K8e and its two closest relatives were created using BLAST2 algorithm (parameters are given in Supplemental material). The RnERV-K8e element (triangle) is plotted on the x-axis; the y-axis represents MmERV-K10c (filled square, left) and Mm_NT_039515.6 (filled circle, right). For each subfamily, the estimated copy number is indicated in brackets. The thicknesses of the diagonal lines are out of scale. Accession numbers are given in the Supplemental material.











