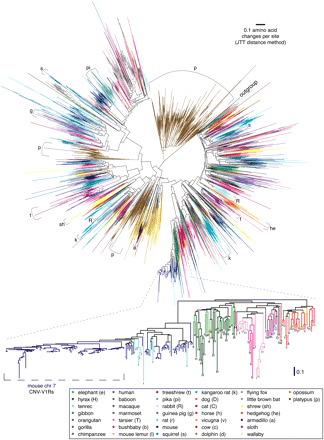
V1R phylogenetic tree showing that species-specific gene duplications have resulted in “semi-private” V1R repertoires. The tree shows the relationships between predicted protein sequences of 4954 of the V1Rs we identified, including intact genes, sequences that contain inactivating mutations/sequence errors (frameshifts corrected where necessary for alignment), and some genes for which full-length sequence is not available, but excluding sequences too short for phylogenetic analysis (see Methods). We also included zebrafish V1R1 (GenBank ABL01523) as an outgroup sequence to root the tree. We used the neighbor-joining method with Jones-Taylor-Thornton amino acid distances to create a tree. Terminal branches are colored according to species. Internal branches share the species color if all descendent terminal taxa are from the same species, or black if descendents are from two or more species. Thus, clusters of same-colored branches reflect species-specific duplications (and possibly some post-speciation gene-conversion events). Internal black branches help distinguish species-specific clades from clades of mixed species especially when the colors used to represent those species are similar. Due to the difficulties of displaying such a large phylogenetic tree and finding enough colors to distinguish so many species, we also provide subtrees constructed from only glires (rodents and lagomorphs) or primate V1Rs in Supplemental Figures 2 and 3. The lower part of the figure provides a higher resolution view of a selected part of the tree and uses letters (see key) to label non-mouse V1Rs to help distinguish species. All species-specific clades containing more than 30 members are indicated in the large tree using brackets with letters to indicate species (see key), except for a clade containing mouse V1Rs that are found in a large copy-number variable (CNV) region (see text), which is instead labeled in the higher-resolution subtree in the lower half of the figure.











