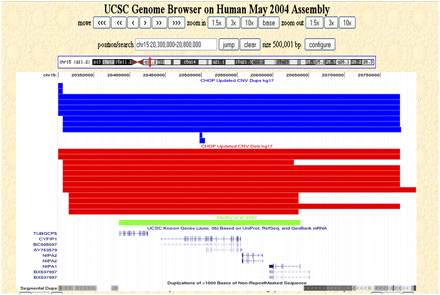
Copy number variation within 15q11.2. Nonunique CNVs detected in our control data set that map within 15q11.2 (chr15:20,300,000–20,800,000, hg17, NCBI build 35) are shown as custom tracks within the UCSC Genome Browser (http://genome.ucsc.edu/). (Red rectangles) Deletions; (blue rectangles) duplications; (green rectangle) and the CNV reported by Murthy et al. (2007). The UCSC known genes and segmental duplication tracks are also shown.











