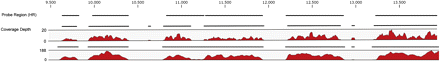
Graphic overview of mapping analysis of an Illumina paired-end sequencing run with a human genomic DNA sample enriched for the genes BRCA1 and TP53. Shown is the capture probe region used for array-based enrichment (black line at top), coverage depth distribution obtained from the first enrichment cycle (middle), and coverage depth distribution from the second enrichment cycle (bottom) to a representative part of the TP53 gene (nucleotides ∼9500–14,000). The obtained consensus sequences are shown as black lines. X-axis, the nucleotide position of the gene; y-axis, the fold coverage depth. Note that the scale of the y-axis varies between the two mappings.











