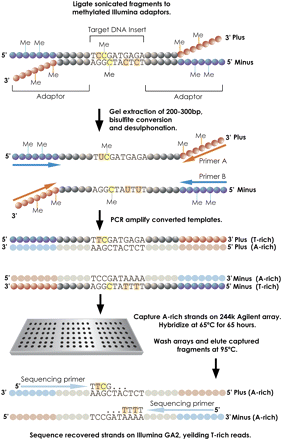
Bisulfite capture procedure. Genomic DNA was randomly fragmented according to the standard Illumina protocol and ligated to custom-synthesized adaptors in which each C was replaced by 5-meC. The ligation was size-fractionated to select material from 150–300 bases in length. The gel-eluted material was treated with sodium bisulfite (see Methods) and then PCR-enriched using Illumina paired-end PCR primers. The resulting products were hybridized to custom-synthesized Agilent 244K arrays containing probes complementary to the A-rich strands. Hybridizations were carried out with Agilent array CGH buffers under standard conditions. After washing, captured fragments were eluted in water at 95°C and amplified again prior to quantification and sequencing on the Illumina GA2 platform.











