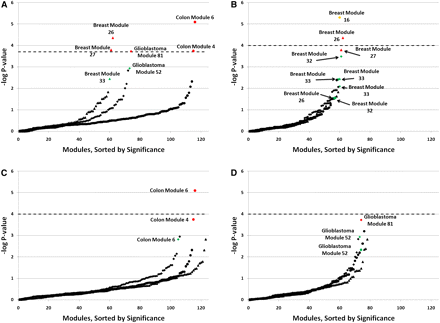
Enrichment of cancer mutations in breast and colorectal modules: The negative log P-value, as determined by the hypergeometric distribution, of the number of mutations mapping to each coexpression module, is plotted in order of significance. The threshold for statistical significance is denoted by the dashed line. (A) Mutations mapped to their corresponding tissue. (B) Breast modules; (C) colon modules; and (D) glioblastoma modules. In each panel, triangles represent modules mapped with breast mutations, circles represent modules mapped with colon mutations, and diamonds represent modules mapped with glioblastoma mutations. Red points are statistically significant for mutations mapped to their corresponding tissue, i.e., above the significance threshold in A. Yellow points are statistically significant for mutations mapped to other tissues. Green points are marginally significant modules of interest.











