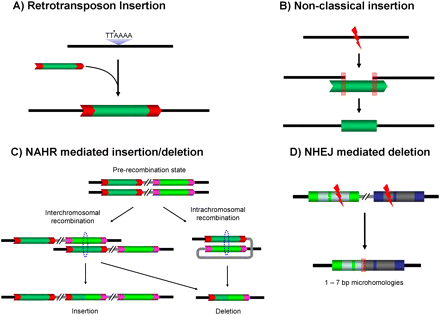
Four types of common MASVs in the HuRef genome. (A) Classical retrotransposon insertion; (B) nonclassical insertions; (C) nonallelic homologous recombination-mediated insertion/deletion; (D) nonhomologous end-joining-mediated deletion. (TTAAAA) Standard L1 cleavage site for classical retrotransposition; (black lines) flanking regions, (gray lines) intervening regions, (dotted circles) homologous recombining regions, (red boxes) microhomology regions, (red arrow boxes) TSDs of each element.











