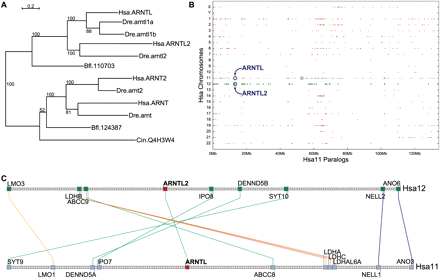
Analysis of the ARNTL gene family. (A) ARNTL phylogenetic tree based on maximum likelihood showing that Danio rerio (Dre) arntl1a is paralogous to arntl1b, and that both of these genes are co-orthologous to human (Hsa) ARNTL. The tree suggests that Dre arntl2 is orthologous to Hsa ARNTL2. (Bfl) Amphioxus; (Cin) Ciona intestinalis. The tree was generated with PhyML (Guindon and Gascuel 2003) using a maximum likelihood algorithm with a GTR model and gamma-distributed rate variation. Bootstrap values are reported on the internal nodes. (B) Human chromosome 11 (Hsa11) paralogy dotplot. Each gene on Hsa11 is represented as a gray dot with its corresponding paralogs plotted as red crosses directly above or below the Hsa11 gene, but shown on the paralog's respective chromosome. ARNTL (Hsa11) and ARNTL2 (Hsa12) are circled. A large region of conserved synteny inhabits the short arm of Hsa11 (the centromere is a gray circle) and Hsa12 (paralogs indicated by green crosses). Other extensive paralogons are on Hsa1 and Hsa19. (C) The ARNTL and ARNTL2 paralogous syntenic cluster in humans is characterized by an inversion of six pairs of genes with ARNTL and ARNTL2 serving as the pivot (50-gene sliding window).











