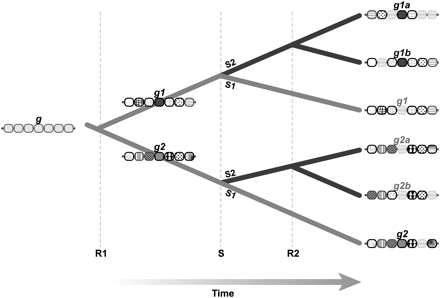
Differential gene loss following whole-genome duplication creates ohnologs gone missing. This image shows the evolutionary history of a gene g and neighbors undergoing a whole-genome duplication event (R1), a speciation event (S), and a second WGD event (R2) that occurs in only one of the descendant lineages, S2. If no changes to the order or composition of genes on the chromosomes occur over time, most algorithms would find that g1a and g1b are co-orthologous to g1, and that g2a and g2b are co-orthologous to g2. In a more realistic evolutionary history, gene losses and chromosomal rearrangements follow the genome duplication event, including a loss of g1 from the S1 lineage and g2a and g2b from the S2 lineage. Due to these losses, orthology assignment algorithms will usually infer that g1a and g1b are co-orthologous to g2, incorrectly assigning co-orthology where there is none. Extinct genes are shown in gray.











