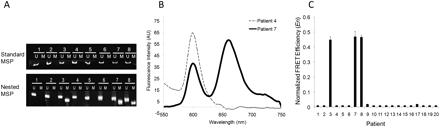
Detection of methylation in human sputum samples. (A) Representative gel from sputum DNA using conventional MSP for PYCARD for eight patients indicates the presence of only unmethylated products. Electrophoresis gel from nested MSP products detects methylation in Patients 3, 7, and 8. (B) Representative fluorescence spectra from two patients with differing methylation status. Significant acceptor (Cy5) emission at 670 nm is observed for patients with methylated PYCARD promoter (yellow trace). (C) Normalized FRET efficiencies (En) for 20 patients, conducted in a blinded fashion, indicate that Patients 3, 7, and 8 have methylation for PYCARD. An arbitrary En cut-off of 0.1 is used to determine positive methylation. All patients show unmethylated PYCARD product as well (data not shown).











