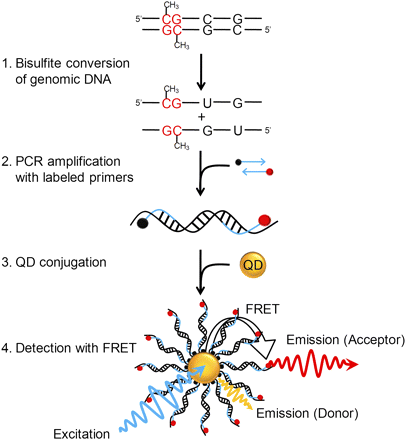
Principle of MS-qFRET for detection of DNA methylation. In step 1, extracted genomic DNA is subject to sodium bisulfite conversion, wherein unmethylated cytosines are converted to uracil while methylated cytosines remain unaffected. In step 2, DNA is amplified using PCR wherein the forward and reverse primers are labeled with a biotin (black dot) and a fluorophore (red dot), respectively. In step 3, the resulting labeled-PCR product is captured by streptavidin functionalized QDs through streptavidin-biotin affinity. Finally, in step 4, upon suitably exciting the QD, the nanoassembly formed allows for FRET to occur between the QD donor and the fluorophore acceptor. Consequently, the labeled-PCR products are detected by emissions of fluorophores accompanied by quenching of QDs to reveal the status of DNA methylation.











