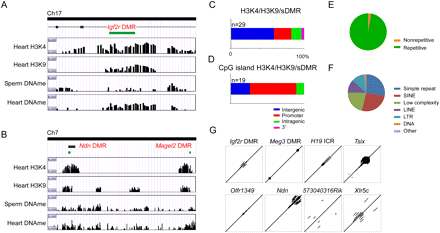
Overlapping patterns of H3K4me3 and H3K9me3 at sDMRs identified imprinting control regions. (A) An example of the Igf2r DMR in intron 2 of the Igf2r locus. The CpG island (green box) indicates the DMR, which contains both H3K4me3 and H3K9me3 modifications and differentially methylated DNA between sperm and heart. (B) Examples of the Ndn and Magel2 DMR showing overlapping H3K4me3 and H3K9me3, as well as sDMR between sperm and heart. (C) Genomic locations of regions with a sDMR and overlapping H3K4me3 and H3K9me3. (D) Genomic locations of CpG islands with a sDMR and overlapping H3K4me3 and H3K9me3. (E) Sequence characterization of regions with a sDMR and overlapping H3K4me3 and H3K9me3 indicated regions are highly repetitive and (F) are comprised mostly of simple repeats, SINES, and low complexity repeats. (G) Examples of sequence alignments for known ICRs (e.g., Igf2r DMR, Meg3 DMR, H19 ICR, and Tsix) and putative ICRs (e.g., Olfr1349, Upstream Ndn, 5730403M16Rik, and Xlr5C) showing repeated elements.











