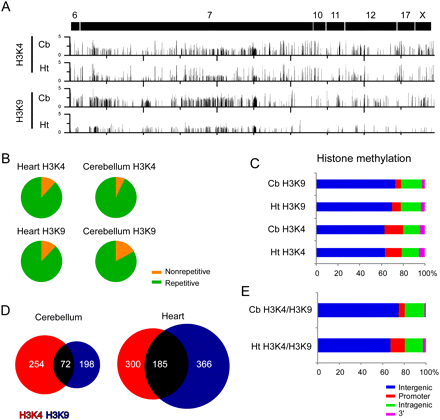
H3K4me3 and H3K9me3 enriched regions at imprinted gene clusters. (A) ChIP-chip profiles of histone modifications for H3K4me3 and H3K9me3 in heart and cerebellum. Y-axis values represent log2 IP/IN. (B) Sequence characterization of regions enriched for H3K4me3 and H3K9me3 showed that each modification was present primarily at repetitive sequences. (C) Genomic location of H3K4me3 and H3K4me3 modifications relative to the RefSeq gene annotation. (D) Venn diagram showing overlapping H3K4me3 and H3K9me3 regions in both heart and cerebellum. (E) Genomic locations of overlapping H3K4me3 and H3K9me3 modifications relative to the RefSeq gene annotations.











