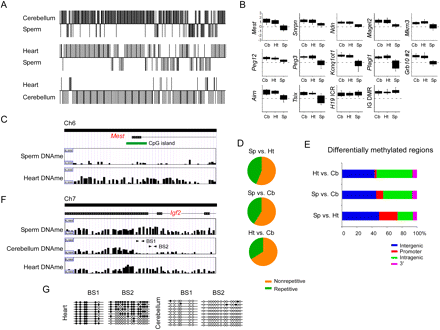
Characterization of sperm-specific and tissue-specific differentially methylated regions at imprinted gene clusters. (A) A sliding window t-test was performed between sperm and heart (Sp vs. Ht; sDMR), sperm and cerebellum (Sp vs. Cb; sDMR), and heart and cerebellum (Ht vs. Cb; tDMR) using the log2 IP/IN ratios obtained from MeDIP to determine sperm and tissue-specific differentially methylated regions (sDMR and tDMR). The y-axis indicates regions that have significantly different (P < 0.00001) methylation profiles between samples and the direction of methylation. (B) The mean log2 IP/IN of probes was calculated over each known DMR in cerebellum, heart, and sperm. Box plots show less methylation in sperm DNA at each DMR, except at the H19 ICR and IG-DMR, which are methylated in sperm and differentially methylated in heart and cerebellum. Box plots represent the 25th and 75th percentile for each averaged sample. Whiskers show 10th and 90th percentiles. (C) An example showing the different patterns observed between sperm DNA methylation (Sperm DNAme) and heart DNA methylation (Heart DNAme) at the Mest DMR. (D) Sequence characterization of sDMR and tDMR showed that nonrepetitive DNA is primarily differentially methylated at imprinted gene clusters. (E) Genomic location of sDMRs and tDMRs relative to the RefSeq gene annotation. (F) An example of a tDMR at the 3′ end of the Igf2 gene. (G) Bisulfite sequencing confirmed loss of methylation in the cerebellum DNA at the 3′ end of the Igf2 gene. Arrows in F indicate regions amplified for bisulfite sequencing.











