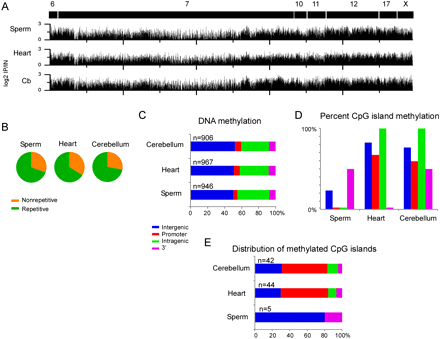
DNA methylation patterns at imprinted gene clusters. (A) MeDIP was performed on DNA isolated from sperm, heart, and cerebellum and the enriched (positive log2 IP/IN) values are plotted for each imprinted gene cluster. Chromosomal regions are marked as black boxes with corresponding chromosome numbers. (B) Sequence characterization of methylated regions in sperm, heart, and cerebellum showed that the imprinted gene clusters were primarily methylated at repetitive DNA based on repeat masker. (C) Plots showing the genomic location of methylated regions relative to the RefSeq gene annotation. (D) CpG island methylation was less frequent in sperm samples compared to the heart and cerebellum, particularly for the promoter and intragenic regions. (E) The majority of methylated CpG islands in heart and cerebellum were present in the promoters of known imprinted genes.











