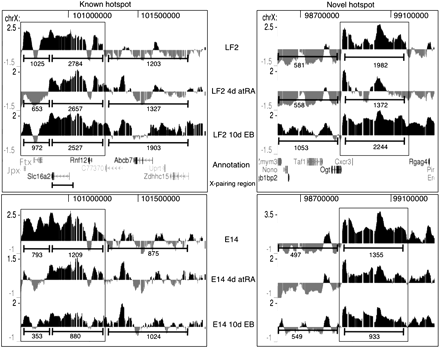
Kinetics of the two H3K27me3 hotspots (which are boxed) surrounding the Xic in LF2 and E14 ES cells as obtained by H3K27me3 ChIP-chip. Screen shots from the UCSC Genome Browser showing the distribution of the H3K27me3 ChIP-chip profiles across a ∼0.5 Mb region of mouse chromosome X in mouse embryonic stem cells (UCSC Mouse [mm9], July 2007; chromosome X genomic coordinates 100.68–101.25 and 98.88–99.31 Mb for the known hotspot and the novel hotspot, respectively). The MM9 coordinates of the Xic are ∼100.3–101 Mb. Annotation is provided below the profiles. For further details, see Figure 1. For quantification using the ChIP-Seq tags of the XT67E1 cells, the “known” hotspot was split into two parts based on their differential kinetics. For comparison, tags were also counted in equally sized neighboring regions.











