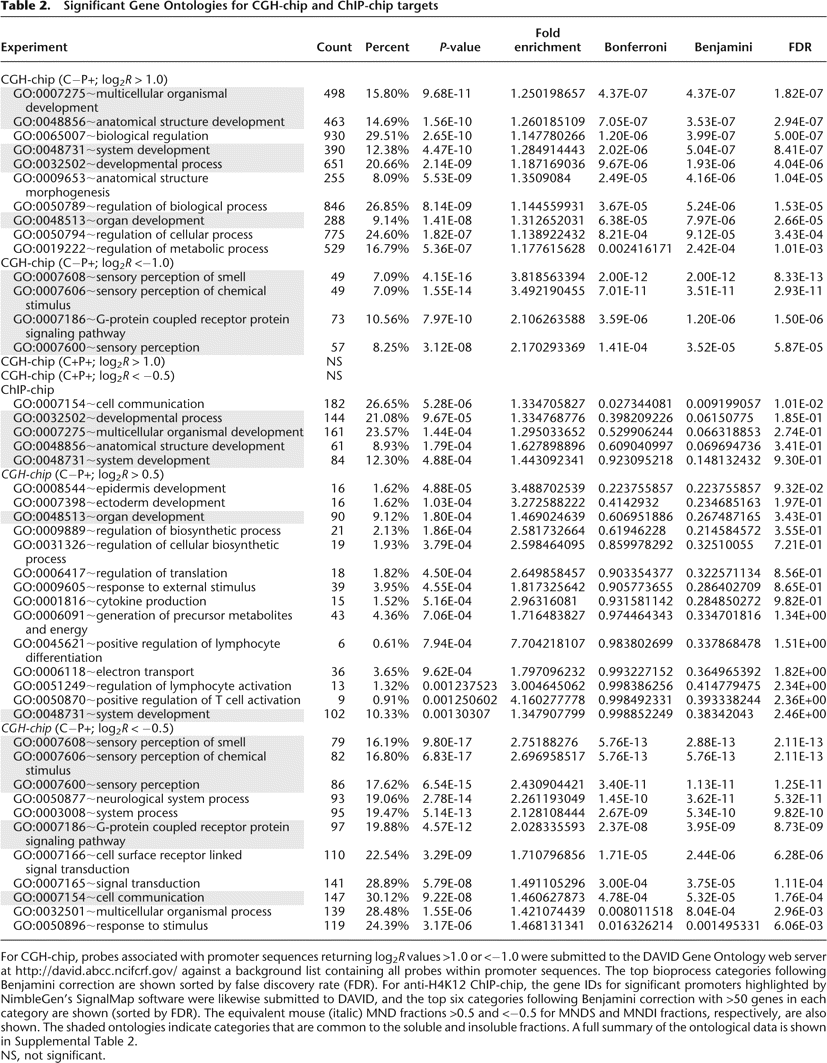
Significant Gene Ontologies for CGH-chip and ChIP-chip targets
Click on table to view larger version.
-
For CGH-chip, probes associated with promoter sequences returning log2R values >1.0 or <−1.0 were submitted to the DAVID Gene Ontology web server at http://david.abcc.ncifcrf.gov/ against a background list containing all probes within promoter sequences. The top bioprocess categories following Benjamini correction are shown sorted by false discovery rate (FDR). For anti-H4K12 ChIP-chip, the gene IDs for significant promoters highlighted by NimbleGen's SignalMap software were likewise submitted to DAVID, and the top six categories following Benjamini correction with >50 genes in each category are shown (sorted by FDR). The equivalent mouse (italic) MND fractions >0.5 and <−0.5 for MNDS and MNDI fractions, respectively, are also shown. The shaded ontologies indicate categories that are common to the soluble and insoluble fractions. A full summary of the ontological data is shown in Supplemental Table 2.
-
NS, not significant.











