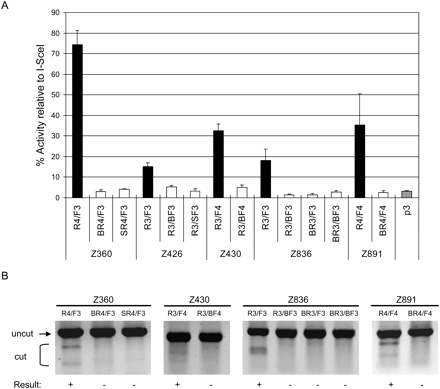
Module swap analysis. New ZFNs were prepared using engineered ZFs to test whether these ZFNs can functionally replace ZFNs composed exclusively of naturally occurring modules that recognize identical sequences. The names of these ZFNs are indicated by the Z numbers at (A) the bottom of the graph or (B) the top of the gel panels. The ZFN monomers whose names start with “B,” such as “BR4,” are composed exclusively of Barbas modules, and those with “S,” such as “SR4,” are composed exclusively of Sangamo modules. The ZFs that compose these ZFNs are described in Supplemental Table 4. These ZFNs were analyzed using (A) the cell-based SSA system and (B) the T7E1 assay. (A) Luciferase activities of cells in which the various ZFNs were expressed. (Gray bar) p3 is the empty plasmid, which was used as a negative control. The target sequence contains the recognition site of I-SceI, which was used as a positive control. The activity of each ZFN pair is reported as the percentage relative to the I-SceI control. ZFN pairs and their constituent monomers are indicated. Means and standard deviations (error bars) from at least three independent experiments are shown. (B) ZFN-mediated genomic modification revealed by the T7E1 assay. ZFN pairs and their constituent monomers are shown at the top of the agarose gels. (+) ZFN pairs that gave rise to positive gene-editing events.











