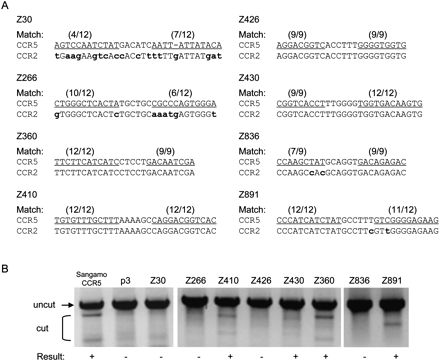
Off-target effects of ZFNs at the CCR2 locus. (A) ZFN recognition elements at the CCR5 and CCR2 loci. The ZFN pairs are indicated at the left of the DNA sequences. (Parentheses) The numbers of base matches between the CCR5 and CCR2 loci; (lowercase bold letters) mismatched bases; (underlined) the half-site ZFN recognition elements. (B) T7E1 assay at CCR2 sites. PCR-amplified DNA corresponding to the CCR2 coding region from cells treated with the ZFN pairs (shown at the top of the gel panels) was analyzed. (+) ZFN pairs that gave rise to the modification at the CCR2 sites. p3 is the empty plasmid used as a negative control. Sangamo's CCR5-targeting ZFN pair (Sangamo CCR5) was included in this assay.











