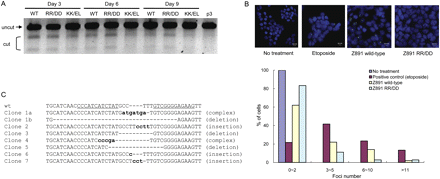
Obligatory heterodimeric ZFNs and isolation of mutant clones. (A) Time-course analysis of wild-type and obligatory heterodimeric ZFNs. The T7E1 mismatch detection assay was performed at various time points with DNA isolated from cells treated with different forms of the FokI nuclease domain. (WT) Wild-type nuclease domain; (RR/DD or KK/EL) obligatory heterodimeric nuclease domains. (B) ZFN-induced DSBs detected by TP53BP1 staining. Both the wild-type and obligatory heterodimeric Z891 ZFN pair were transfected into HEK293T/17 cells, and intracellular TP53BP1 foci were detected by immunofluorescence at day 2 post-transfection. Etoposide (1 μM) was used as a positive control. The distribution of the numbers of TP53BP1 foci is plotted. At least 100 cells were analyzed for each treatment in two independent measurements. (C) DNA sequences of mutant clones. Seven mutant clones were isolated, by limiting dilution, from cells treated with the RR/DD ZFN dimer. The DNA sequences of the target site in these clonal cells are shown. (Dashes) Deletions are indicated with dashes; (small letters) inserted bases. Clones 1a and 1b indicate DNA sequences that resulted from biallelic modifications in a single clone.











