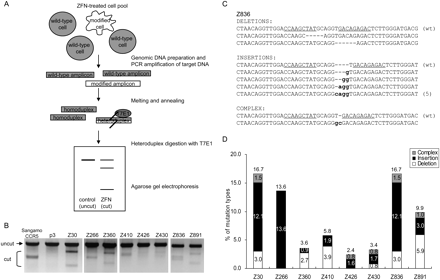
ZFN-mediated genome editing in human cells. (A) Schematic overview of mismatch detection using T7E1 assay. Genomic DNA was purified from cells transfected with plasmids encoding ZFNs. The DNA segments encompassing the sites of ZFN recognition were PCR-amplified, and the DNA amplicons were melted and annealed. If the DNA amplicons contain both wild-type and mutated DNA sequences, heteroduplexes would be formed. T7E1 recognizes and cleaves heteroduplexes, but not homoduplexes. The DNA fragments were assessed by agarose gel electrophoresis; a schematic of an idealized gel result is shown. (B) ZFN-mediated genomic modification revealed by the T7E1 assay. The ZFN pairs are shown at the top of the agarose gels. The expected positions of the resulting DNA bands are indicated by an arrow (uncut) and a bracket (cut) at the left of the gel panels. p3 is the empty plasmid used as a negative control. Sangamo's CCR5-targeting ZFN pair (Sangamo CCR5) was included in this assay. (C) DNA sequences of a genomic site targeted by the Z836 ZFN pair. (Underlined) ZFN recognition elements. (Dashes) Deletions; (small letters) inserted bases. In cases in which a mutation was detected more than once, the number of occurrences is shown in parentheses. (wt) Wild type. (D) Types of mutations at various ZFN-targeted sites. The number of deletions, insertions, and complex mutations for each ZFN pair (as shown in C for Z836) were counted, and the percentages of these incidents were plotted.











