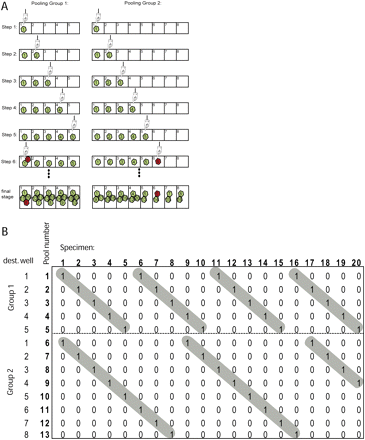
An example of pooling design. (A) The steps involved in pooling 20 specimens with two pooling patterns: (left) n ≡ r2 (mod 5); (right) n ≡ r2 (mod 8). Note that each pattern corresponds to a pooling group and to a set of destination wells. For simplicity, the destination wells are labeled 1–5 and 1–8 instead of 0–4 and 0–7. In this example, we choose specimen #6 to be mutant (red, green = wild type) among the 20 specimens. (B) The corresponding pooling matrix. The matrix is 13 × 20 and partitioned into two regions (broken line) that correspond to the two pooling patterns. The (highlighted in gray) staircase pattern in each region is typically created in our pooling scheme. The weight of the matrix is 2, the maximal intersection between each set of two column vectors is 1, and the maximal compression rate is 4.











