Mapping of the 3′-polyadenylation sites of a panel of selected genes
Click on table to view larger version.
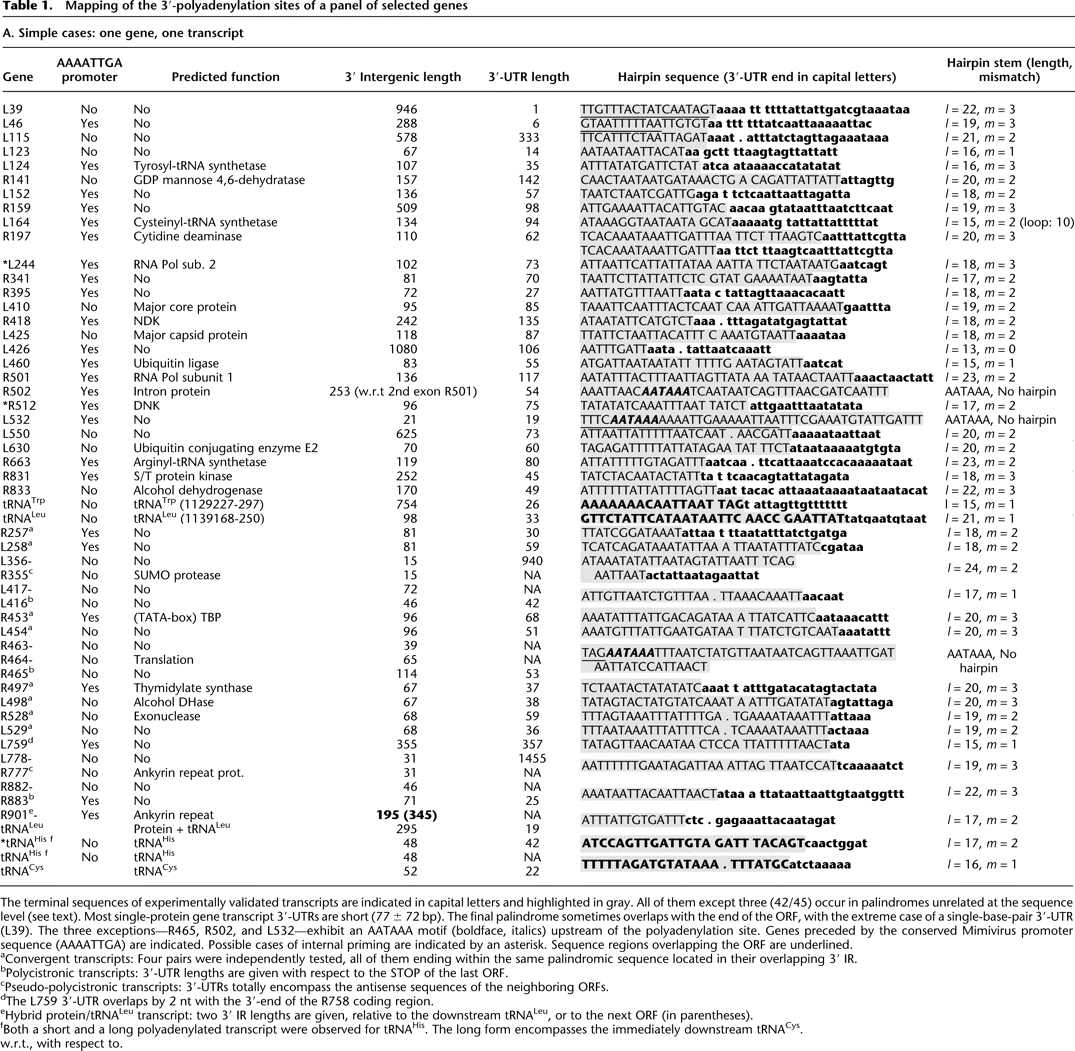
-
The terminal sequences of experimentally validated transcripts are indicated in capital letters and highlighted in gray. All of them except three (42/45) occur in palindromes unrelated at the sequence level (see text). Most single-protein gene transcript 3′-UTRs are short (77 ± 72 bp). The final palindrome sometimes overlaps with the end of the ORF, with the extreme case of a single-base-pair 3′-UTR (L39). The three exceptions—R465, R502, and L532—exhibit an AATAAA motif (boldface, italics) upstream of the polyadenylation site. Genes preceded by the conserved Mimivirus promoter sequence (AAAATTGA) are indicated. Possible cases of internal priming are indicated by an asterisk. Sequence regions overlapping the ORF are underlined.
-
aConvergent transcripts: Four pairs were independently tested, all of them ending within the same palindromic sequence located in their overlapping 3′ IR.
-
bPolycistronic transcripts: 3′-UTR lengths are given with respect to the STOP of the last ORF.
-
cPseudo-polycistronic transcripts: 3′-UTRs totally encompass the antisense sequences of the neighboring ORFs.
-
dThe L759 3′-UTR overlaps by 2 nt with the 3′-end of the R758 coding region.
-
eHybrid protein/tRNALeu transcript: two 3′ IR lengths are given, relative to the downstream tRNALeu, or to the next ORF (in parentheses).
-
fBoth a short and a long polyadenylated transcript were observed for tRNAHis. The long form encompasses the immediately downstream tRNACys.
-
w.r.t., with respect to.











