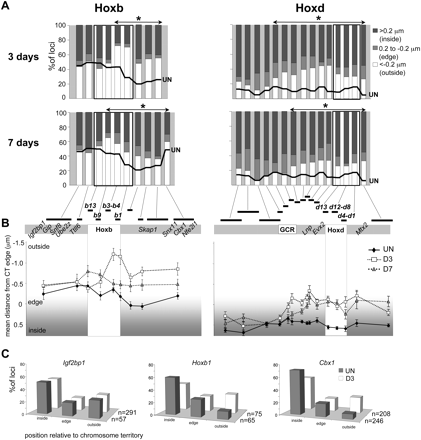
Chromosome territory reorganization during ES cell differentiation. (A) Histograms showing the percentage of signals across the Hoxb or Hoxd regions located on either inside (>0.2 μm; dark gray bars), at the edge (±0.2 μm; light gray bars), or outside (<−0.2 μm white bars) of the MMU11 (Hoxb) or the MMU2 CT (Hoxd), as measured by 2D FISH in OS25 ES cells differentiated for 3 or 7 d. The thick black line shows the corresponding data in undifferentiated ES cells (from Fig. 2B). Probes showing a further significant (P < 0.05) relocalization toward the outside of the CT during differentiation are indicated by the asterisked regions (n = 100). (B) Mean position (μm) ± SEM, measured by 2D FISH, of the Hoxb genomic region relative to the edge of MMU11 CT (left), or of the Hoxd genomic region relative to the edge of MMU2 CT (right) in undifferentiated OS25 ES cells (filled diamonds) and in cells differentiated for 3 d (open squares) or 7 d (shaded triangles). (C) Position of Hoxb1 and 5′ (Ifg2bp1) or 3′ (Cbx1) flanking regions relative to the MMU11 CT (inside of, edge, or outside of) assayed by 3D FISH in pFa fixed undifferentiated ES cells (filled bars), and cells differentiated for 3 d (open bars).











