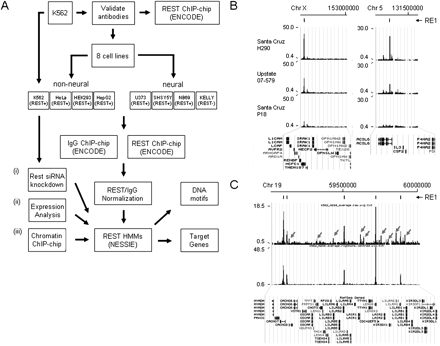
Approach to identify REST binding sites across the ENCODE regions. (A) Initially, three anti-REST antibodies were validated for enrichment levels in K562 cell lines (see also panel B). One of these, as well as rabbit IgG antisera, was used for subsequent ChIP-chip across the ENCODE regions in eight diverse human cell lines representing both neural and non-neural lineages. REST datasets were normalized with rabbit IgG datasets and resultant peaks of enrichments were identified computationally using a hidden Markov model (NESSIE). RE1 DNA motifs and target genes found at or near various classes of REST binding sites were subsequently identified. In parallel, K562 cells were also used to generate expression data and ChIP-chip data for chromatin states (histone modifications, FAIRE, and histone density) and to perform siRNA experiments against REST. (B) UCSC genome browser visualization of enrichments levels obtained in K562 cells using three anti-REST antibodies across two ENCODE regions (ENm006 and ENm002) upstream of the L1CAM (left) and ACSL6 (right) genes, respectively. (Top) Genome coordinates. Enrichment scales (y-axis) in both regions are the same for all three antibodies. Performance of antibody H290 was in the range of ∼2–10 times higher than the other two antibodies tested. (C) (Top panel) UCSC genome browser view of ChIP-chip enrichments (y-axis) detected across the 1-Mb ENCODE region ENm007 (genome coordinates along x-axis shown at top of panel) for raw anti-REST antibody-derived data. (Lower panel) Same data that have been normalized using data derived from the corresponding rabbit IgG dataset. (Black bars, top) The five resultant “normalized” peaks all are associated with canonical RE1 motifs (Johnson et al. 2006); (gray arrowheads) peaks of REST enrichment that were removed when correcting the data in this way.











