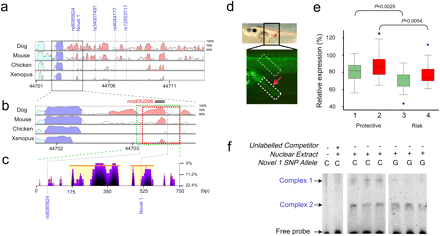
(a) VISTA view of the occurrence of conserved sequence domains in the SMAD7 risk-associated region. Shown from top to bottom are global alignments of human vs. dog, mouse, chicken, and Xenopus tropicalis, respectively. Colored peaks indicate regions of at least 100 bp in length and with 75% sequence similarity. Cyan peaks are UTR, purple peaks are coding regions, and pink peaks are noncoding regions. (b) Detail of the genomic regions used in the Xenopus functional enhancer assays (green and red blocks). Note that this region contains an evolutionarily-conserved noncoding peak. (c) eShadow view (ClustalW multiple sequence alignments presented as percent mismatch) of the SMAD7 construct showing regions of sequence conservation amongst primates (Human, Chimp, Orangutan, Rhesus, and Marmoset). Note that SNP Novel 1 is conserved in primates. (d) The tested regions contain an enhancer that promotes reporter gene expression in the rectal region of Xenopus tadpoles. The bright field image above shows a 5-d tadpole embryo. The rectal region is indicated by a red arrow. The fluorescent image below shows a detail of the rectal region of a Xenopus transgenic embryo in which GFP expression is promoted by the enhancer. The intensity of the rectal expression promoted by the enhancer from the Protective or the Risk haplotypes was measured relative to the signal observed in a fixed area in the muscles region (boxed in gray), which was considered as 100%. The DNA tested contains either the protective or risk variants of both rs8085824 and Novel 1 (green; 1 and 3) or solely Novel 1 (red; 2 and 4). (e) Box-whisker plot of the relative expression observed in transgenic embryos harboring the Protective or the Risk DNA promoting GFP expression. The enhancer from the risk haplotype/allele shows a significantly decreased enhancer activity. (f) EMSA revealing allele-specific binding of unknown nuclear factors at Novel 1 SNP.











