Coverage and accuracy of the Illumina 1M BeadChip genotyped sites of the called consensus sequence
Click on table to view larger version.
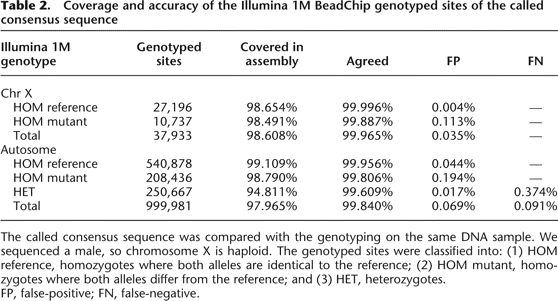
-
The called consensus sequence was compared with the genotyping on the same DNA sample. We sequenced a male, so chromosome X is haploid. The genotyped sites were classified into: (1) HOM reference, homozygotes where both alleles are identical to the reference; (2) HOM mutant, homozygotes where both alleles differ from the reference; and (3) HET, heterozygotes.
-
FP, false-positive; FN, false-negative.











