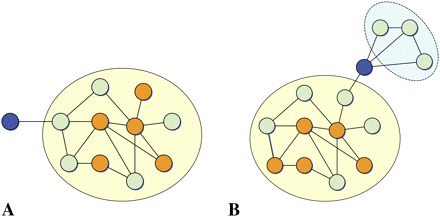
PEXA pruning step. Two types of “superficial” nodes were identified for pruning: (A) nodes that are not siRNA hits and that have only one connection to the network via an interaction with another non-siRNA hit gene, and (B) nodes that are part of a network component comprised solely of non-siRNA hit genes and connected to the network via a non-siRNA hit gene. In the example networks depicted, blue nodes represent the superficial nodes, orange nodes represent siRNA hits, and green nodes represent non-siRNA hit nodes incorporated from either the KEGG database or PPI data. The superficial nodes and subnetworks containing no siRNA hits to which they connect are removed as part of the pruning process.











