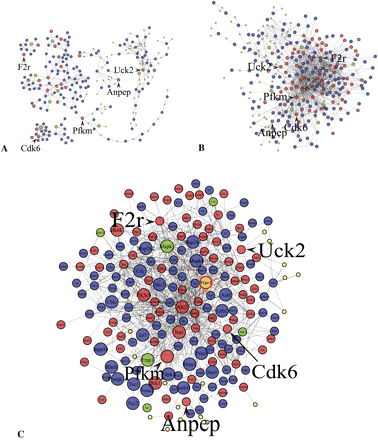
Using PEXA to construct the insulin resistance networks. Red nodes represent genes in the siRNA hit list, green nodes represent genes that were screened but that are not in the siRNA hit list, and blue nodes represent genes that were not screened. (A) The siRNA hits (red nodes) serve as seeds for building up the seeding paths based on pathways represented in the KEGG database. (B) The seeding paths depicted in A are expanded and joined together using PPI data to form a single network. (C) After pruning we obtain a core network of genes enriched for siRNA hits. Larger sized nodes represent genes in the KEGG insulin signaling pathway, while the smaller sized yellow nodes represent small molecules in the KEGG database. The red edges represent interactions between S1pr2 (gold node) and its neighbors. We arbitrarily selected a few nodes and labeled them using a large font size to indicate the interdependency among the three plotted networks.











